A team led by Sandia National Laboratory were able to resolve the interparticle distance of 3D DNA-directed gold nanoparticle assemblies using correlative transmission electron microscopy and fluorescence spectroscopy in liquid. The researchers used Liquid cell TEM platform to evaluate the DNA linkage lengths assembled over a period of time. The results are published in Micron.
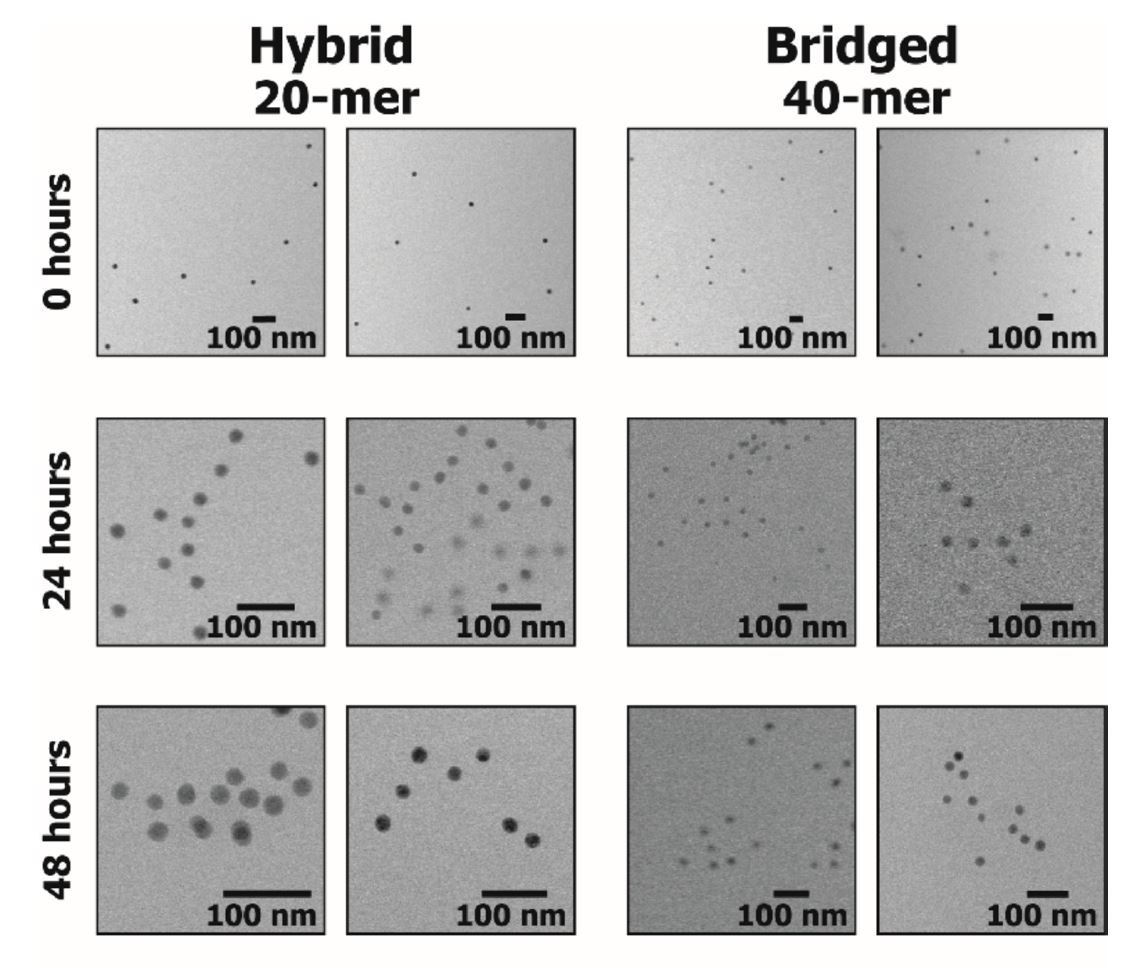
The linkage lengths of 3D assembled DNA architectures are difficult to resolve using bulk characterization techniques, including small angle x-ray scattering. The researchers used liquid cell transmission electron microscopy (TEM) with complementary fluorescence spectroscopy and observed that the architectures of DNA-functionalized Au particles varied from hybrid to bridge depending on the time taken for the assembly as well as the length of the functionalization. This nanoscale imaging technique in solution can be useful tool to optimize other 3D nanoparticle architectures.
Reference: Katherine L. Jungjohann, David R. Wheeler, Ronen Polsky, Susan M. Brozik, James A. Brozik, and Angela R.Rudolph. “Liquid-Cell Scanning Transmission Electron Microscopy and Fluorescence Correlation Spectroscopy of DNA-Directed Gold Nanoparticle Assemblies,” Micron (2018). DOI:10.1016/j.micron.2018.11.004
View All News

